Binary and Multiclass Outcomes
Source:vignettes/articles/Binary-and-Multiclass-Outcomes.Rmd
Binary-and-Multiclass-Outcomes.Rmd
library(nadir)
#> Registered S3 method overwritten by 'future':
#> method from
#> all.equal.connection parallellyBinary Outcomes.
For an example, we show using the Boston dataset and creating a
binary outcome for a regression problem, and then train a
super_learner() to predict this binary outcome.
To handle binary outcomes, we need to adjust the method for
determining weights. This is because we don’t want to use the default
mse() loss function, but instead we should to rely on using
the negative log likelihood loss on the held-out data. To do this
appropriately in the context of binary data, we want to make sure the
loss function used is the
determine_weights_for_binary_outcomes() function provided
by nadir. We can do this either by setting
outcome_type = 'binary' or by passing the function directly
to super_learner() as the argument
determine_super_learner_weights = determine_weights_for_binary_outcomes.
data('Boston', package = 'MASS')
# create a binary outcome to predict
Boston$high_crime <- as.integer(Boston$crim > mean(Boston$crim))
data <- Boston |> dplyr::select(-crim)
# train a super learner on a binary outcome
trained_binary_super_learner <- super_learner(
data = data,
formula = high_crime ~ nox + rm + age + tax + ptratio,
learners = list(
logistic = lnr_logistic, # the same as a lnr_glm with extra_learner_args
# set to list(family = binomial(link = 'logit'))
# for that learner
rf = lnr_rf_binary, # random forest
lm = lnr_lm), # linear probability model
outcome_type = 'binary',
verbose = TRUE
)
# let's take a look at the learned weights
trained_binary_super_learner$learner_weights
#> logistic rf lm
#> 1.000000e+00 2.691182e-16 4.748858e-20
# what are the predictions? you can think of them as \hat{P}(Y = 1 | X).
# i.e., predictions of P(Y = 1) given X where Y and X are the left & right hand
# side of your regression formula(s)
head(trained_binary_super_learner$sl_predict(data))
#> 1 2 3 4 5 6
#> 2.772297e-05 1.034418e-05 5.749397e-06 2.472188e-06 3.237557e-06 3.784687e-06
# classification table
data.frame(
truth = data$high_crim,
prediction = round(trained_binary_super_learner$sl_predict(data))) |>
dplyr::group_by(truth, prediction) |>
dplyr::count() |>
ggplot2::ggplot(mapping = ggplot2::aes(y = truth, x = prediction, fill = n, label = n)) +
ggplot2::geom_tile() +
ggplot2::geom_label(fill = 'white', alpha = .7) +
ggplot2::scale_fill_distiller(palette = 'Oranges', direction = 1) +
ggplot2::scale_x_continuous(breaks = c(0, 1)) +
ggplot2::scale_y_continuous(breaks = c(0, 1)) +
ggplot2::xlab("super_learner() prediction") +
ggplot2::theme_minimal() +
ggplot2::theme(legend.position = 'none') +
ggplot2::ggtitle("Classification Table")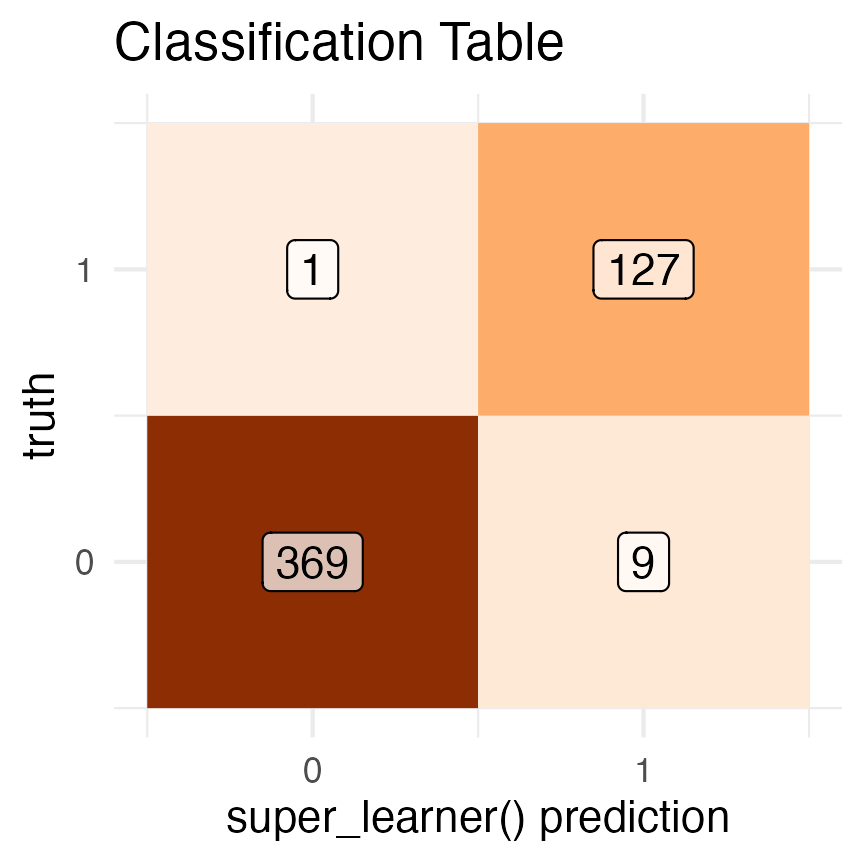
data.frame(
truth = data$high_crim,
predicted_pr_of_1 = trained_binary_super_learner$sl_predict(data)) |>
ggplot2::ggplot(mapping = ggplot2::aes(x = predicted_pr_of_1)) +
ggplot2::geom_histogram() +
ggplot2::facet_grid(truth ~ ., labeller = ggplot2::labeller(truth = ~ paste0('truth: ', .))) +
ggplot2::theme_bw() +
ggplot2::theme(panel.grid.minor = ggplot2::element_blank()) +
ggplot2::xlab(bquote(paste("super_learner() predictions, ", hat(bold(P)), '(Y = 1)'))) +
ggplot2::ggtitle("Classification Task")
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.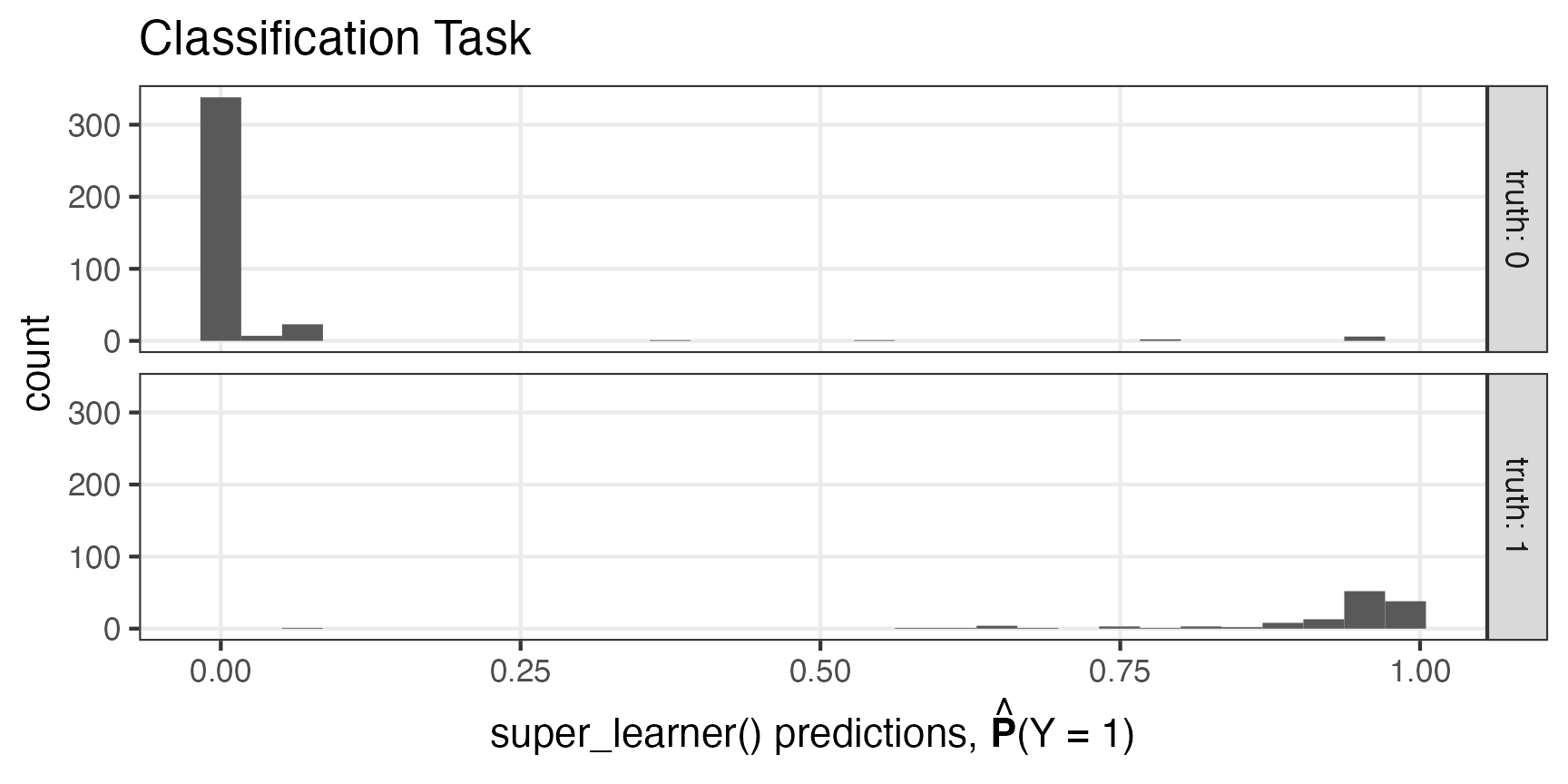
An important thing to know about constructing learners for the binary
outcome context is that
determine_weights_for_binary_outcomes() requires that the
outputs of the learners on newdata are predictions for the
outcome being equal to 1.
Multiclass Regression, i.e., Multinomial Regression
Above we covered binary classification — now we turn to classification problems where the dependent variable is one of a discrete number of unique levels.
nadir includes two learners that are designed for such
multiclass regression problems: lnr_multinomial_vglm and
lnr_multinomial_nnet.
We can perform super learning with them and a classification problem
like that of classifying the penguins’ species in the
palmerpenguins dataset.
library(palmerpenguins)
df <- penguins[complete.cases(penguins),]
sl_learned_model <- super_learner(
data = df,
formulas = list(
.default = species ~ flipper_length_mm + bill_depth_mm,
nnet2 = species ~ poly(flipper_length_mm, 2) + poly(bill_depth_mm, 2) + body_mass_g,
nnet3 = species ~ flipper_length_mm * bill_depth_mm + island
),
learners = list(
nnet1 = lnr_multinomial_nnet,
nnet2 = lnr_multinomial_nnet,
nnet3 = lnr_multinomial_nnet,
vglm = lnr_multinomial_vglm
),
outcome_type = 'multiclass',
verbose = TRUE)
compare_learners(sl_learned_model, loss_metric = negative_log_loss)
#> # A tibble: 1 × 4
#> nnet1 nnet2 nnet3 vglm
#> <dbl> <dbl> <dbl> <dbl>
#> 1 120. 119. 156. 120.
round(sl_learned_model$learner_weights, 3)
#> nnet1 nnet2 nnet3 vglm
#> 0.000 0.000 0.956 0.044